

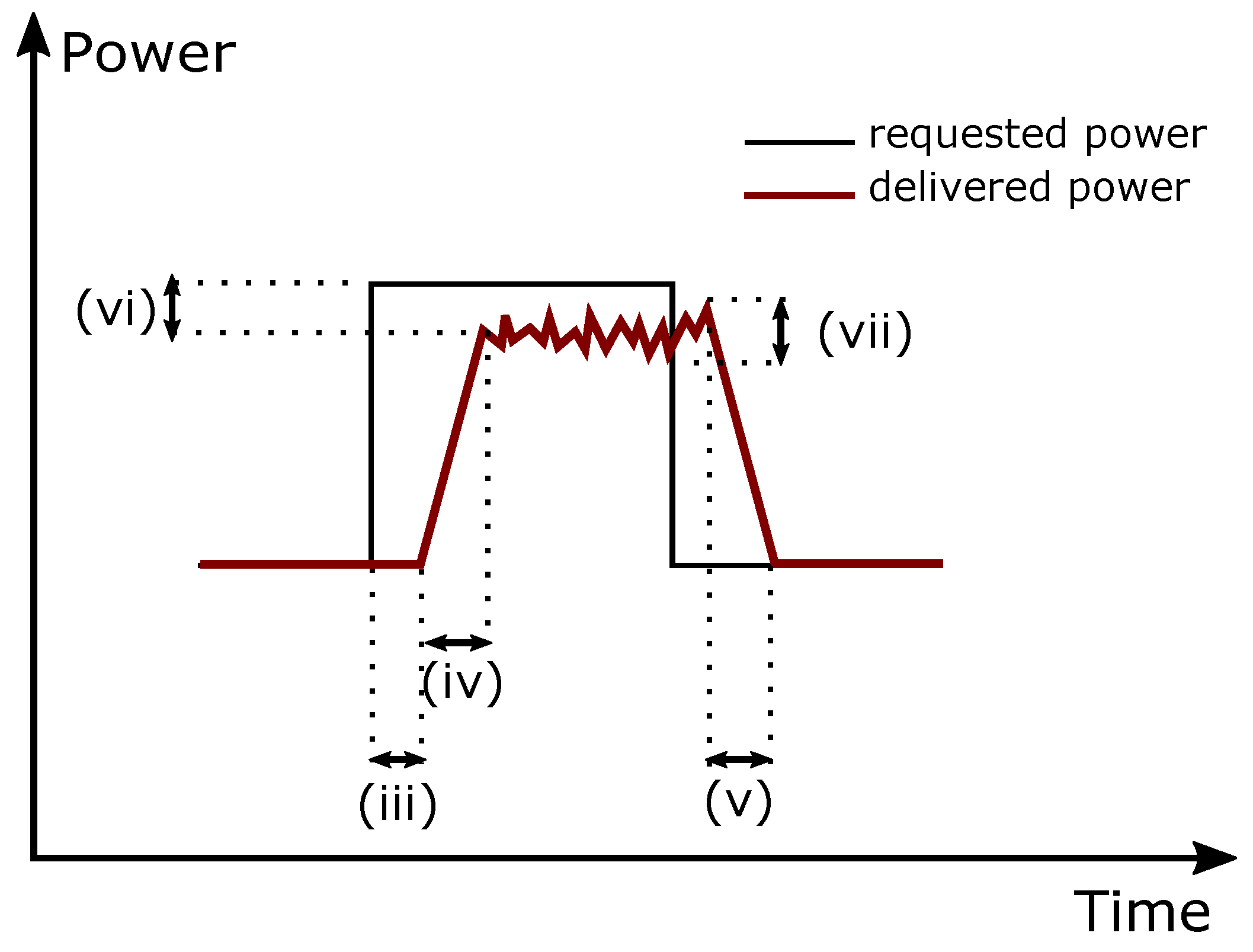
Our study uncovers previously obscured finer-scale genome organization, establishing functional links between chromatin folding and gene regulation. The model receives as input the desired power set-point PowerRequested and provides the signal PowerProvided SIM, similarly to the real operation cases. A schematic representation is shown in Figure 11. Acute inhibition of transcription disrupts these gene-related folding features without altering higher-order chromatin structures. The EV charger model is designed using the Simulink platform within the Matlab MathWorks 2018b software environment. It can be operationally defined as individuals. E-P stripes extending from the edge of domains predominantly link co-expressed loci, often in the absence of CTCF and cohesin occupancy. Neuroticism is an important personal trait and includes in all major modal of personality, such as three-factor 1,2 and five-factor models 3,4,5. All experiments have been run on a computer system with an NVIDIA GeForce 920 M GPU, an Intel® Core i57200 U 2.50 GHz CPU, and 6 GB RAM. Combinatorial binding of transcription factors, cofactors, and chromatin modifiers spatially segregates TAD regions into various finer-scale structures with distinct regulatory features including stripes, dots, and domains linking promoters-to-promoters (P-P) or enhancers-to-promoters (E-P) and bundle contacts between Polycomb regions. In this study, as it is seen in Table 2, Matlab 2018b has been used for performing experiments.
Here, we overcome some limitations of conventional 3C-based methods by using high-resolution Micro-C to probe links between 3D genome organization and transcriptional regulation in mouse stem cells. Whereas folding of genomes at the large scale of epigenomic compartments and topologically associating domains (TADs) is now relatively well understood, how chromatin is folded at finer scales remains largely unexplored in mammals.


 0 kommentar(er)
0 kommentar(er)
